Using scanning electron microscopy and Hitachi map 3D software based on Mountains® technology, cell biology scientists at the University of Miyazaki (Japan) defined a new method for examining stem cell architecture. Their findings were recently published in the academic journal Nature.
A new method for investigating stem cells
Recent advances in biomedical research, such as the production of regenerative organs from stem cells, require three-dimensional analysis of cell and tissue architectures.
High-resolution imaging by electron microscopy is understood to be the best way to elucidate complex cell/tissue architectures, but this method requires very laborious and time-consuming sample preparation.
A team of cell biologists at the University of Miyazaki in Japan, led by Akira Sawaguchi recently presented a new three-dimensional method for assessing cell/tissue structures. This involves embedding cells in paraffin and visualizing 30-μm-thick paraffin sections using backscattered electron imaging in a low-vacuum scanning electron microscope.
Applications in oncological experiments
This new method offered a new wealth of insight in an experiment on pancreatic tumor cells. The so-called “three-dimensional culture system” helped scientists confirm the similarity between xenografts (cells transplanted into mice) and three-dimensionally cultured cells (figures a-e).
Cells were imaged using a scanning electron microscope, equipped with a four-quadrant detector (figure f). These images were processed using Hitachi map 3D software which allowed the construction of a 3D topographic model (figure g). The microstructure, composed of vacuoles (enclosed compartments) containing necrotic cell debris, could be clearly visualized.
It is thought that this technique is applicable in many other domains of medical research and could even enable scientists to re-examine valuable samples prepared decades ago.
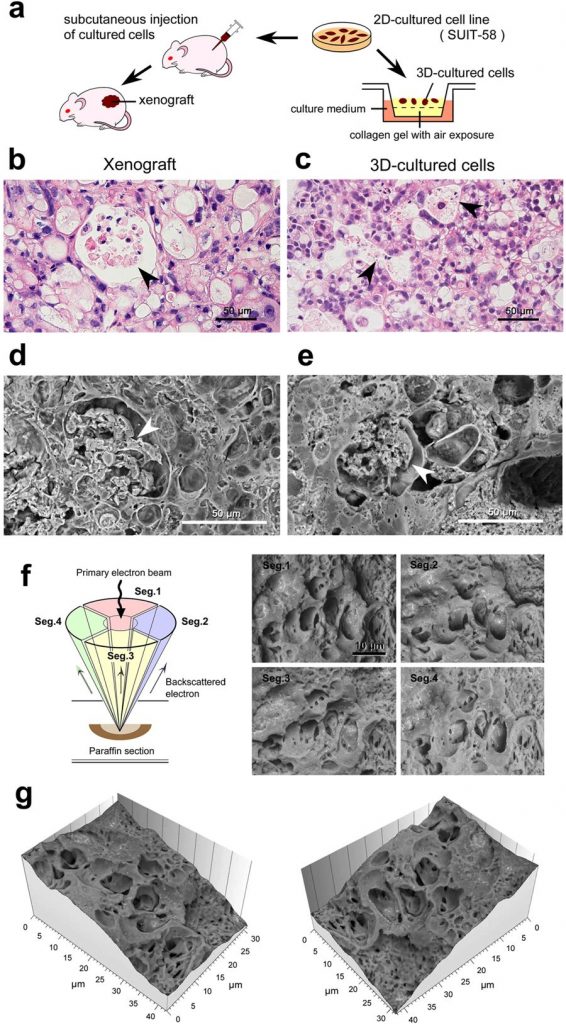
Above: application of the new method for oncological experiments using SUIT-58 cell line. (a) Illustration of the comparative experiments between xenograft and three-dimensionally cultured cells. (b,c) Light micrographs of H&E stained 5 μm sections and (d–g) Thick PS-LvSEM micrographs of 30 μm sections. Note the similar profiles of vacuole formation enclosing necrotic cell debris (arrowheads). (f) Illustration of the four-segment BSE detectors (left) and collected micrographs by each detector (right). (g) Topographic images reconstructed from the raw micrographs of the four-segment BSE images.
Read full article in Nature: doi.org/10.1038/s41598-018-25840-8